Research
Gene network for disease and drug action
When a machine is not working, you want to fix it. To do this properly, you need to know how the machine works, or even better, have a blue print of how it is built. This would allow you to find the underlying defects and devise strategies to repair the machine. When we want to find new ways to treat a retinal-degenerative disease, we also need a blue print—the disease-causing gene network. This network tells us how the molecular components in our body work together, and how a disease causes problems that ultimately affect our vision. If we know these details well, we may find a component in the molecular machine that should be targeted by new drugs to treat retinal degeneration. To this end, we study the disease-causing gene network in zebrafish eye-disease models. These models are prolific and their eyes shared many similarities with humans. Studying the network these models may reveal novel targets for treating patients.
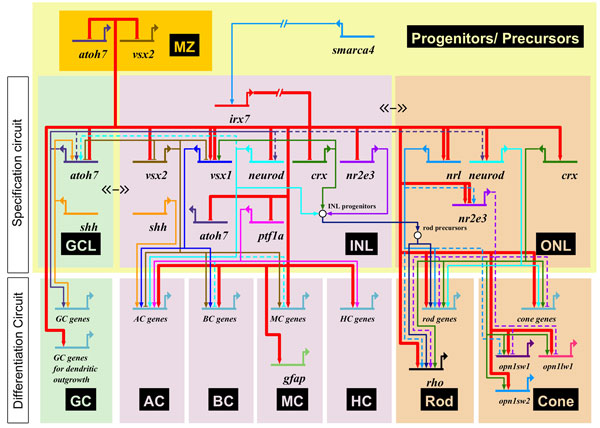 A gene regulatory network for retinal dystrophy. Adapted from Zhang et al., 2012.
A gene regulatory network for retinal dystrophy. Adapted from Zhang et al., 2012.
The first step to elucidate the network is to detect the molecular components that are perturbed by retinal-degenerative diseases. This detection can be specifically done on the microdissected eye tissues, including retina (Leung & Dowling 2005; Zhang & Leung 2010) and retinal pigment epithelium (Leung et al., 2007; Zhang & Leung 2010). These tissues are collected from critical stages of embryonic development during which retinal degeneration occurs. Then, the normal tissues and the disease tissues are compared to reveal the changes in global gene expression. The differences in this comparison indicates the components that are affected in the molecular machine by retinal degeneration.
Microdissection of zebrafish retina. Adapted from Zhang & Leung 2010.









